RESEARCH | Haruko Takeyama Laboratory | Biomolecular Engineering Laboratory, Waseda University
Single cell Research of microbiome using single-cell genetic analysis method
Microorganisms exist everywhere. Since the most microorganisms in the environment are unculturable, there is a need for detailed analysis on genomic information in order to predict each one of their metabolisms. Conventional genetic analysis method for environmental microorganisms contained a mixture of various microbial genomes, therefore, it has been difficult to extract and reconstruct the genome information of an interesting microorganisms from other billions of microbial information.
The single-cell genetic analysis method we are developing can decode the whole genome sequence information from only a single microbial cell with high accuracy. Using this technology, it is possible to acquire high-throughput microbial genome data. By making full use of this method and bioinformatics analysis, We aim to understand the relationship between bacteria communities and the environment・diseases from unknown microbial genome data.
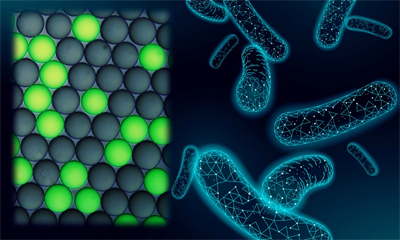
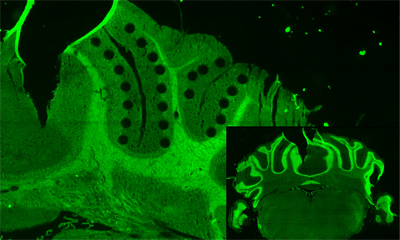
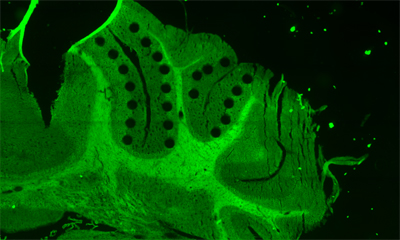
RNA-seq Integrated analysis to understand the tissue in single-cell level
The smallest unit of life is a cell. In order to understand the complex phenomena of life, it is effective to understand that the genes that work in each cell. The remarkable development of DNA sequencing technology has made it possible to conduct comprehensive gene expression analysis. On the other hand, it has become clear that spatial information of cells in tissues is essential for understanding intercellular networks and elucidating biological functions and pathogenesis.
In our laboratory, we are developing technology to extract micro regions from tissue sections (HARD), to analyze DNA and RNA sequences in single-cell or dozens cells (WET), and to analyze spatial tissue structure by integrating the prediction of cells function and the information of tissue morphology (DRY), with the aim of constructing a total system. We are collaborating with various institutions and hospitals to apply our cutting-edge technologies in the aim to understand the functions of living tissue and medical field.
Marine biotechnology Marine biotechnology for Marine environmental bacteria
Ocean, which accounts for seventy percent of the earth’s surface, is considered as an essential resource for microorganisms. Our laboratory excels in the analysis of symbiont microorganisms with marine organisms. Currently, we mainly deal with corals in Okinawa and sponges along Pacific coast (Izu Peninsula, Hachijo-jima island, Miyako-jima island).
In the research using corals, we aim to estimate interaction between microorganisms and hosts, and roles of coexisting bacteria in environments, by carrying out large-scale environmental omics analysis and single-cell genome analysis of coral co-existing microorganisms simultaneously.
In the research using sponges, we aim to discover and characterize sponge-associated microorganisms that produce useful bioactive substances, by combining single-cell genome analysis technology and micro-raman spectroscopy.
We work on advanced researches to acquire new biological knowledge, by combining various technologies our laboratory developed.

Raman spectroscopy Development of technology to search for intracellular molecules by Raman spectroscopy
Raman scattering phenomena was first discovered by Dr. C.V. Raman from India in 1928 and the technology of Raman spectroscopy now has been widely used to identify molecules based on its specific vibrational frequency due to chemical bonding. When applied on fresh samples such as living cells or bacteria, Raman spectroscopy can determine substances at molecular level without any damages or staining process on the samples. Recent advances in hardware and spectral method achieved to give in-real-time images of cellular molecular distributions. Our laboratory is developing a label-free technology to detect various target molecules in living cells and microorganisms, while developing devices suitable for cell measurement and constructing analysis algorithms. We are also working on the development of techniques which enable us to analyze and screen, on the spot and at the single-cell level, the extent of production and the origin of secondary metabolites that are highly dependent on the growth environment, by changing the spectrum of secondary metabolites produced by microorganisms to the form of libraries.

Analysis of information Development of tools and algorithms for analyzing complex data
With the advance of Next-generation sequencing (NGS) technology, the data around biology has become diverse. Our laboratory is developing tools and algorithms for comprehensively analyzing single-cell genome data or spatial transcriptome data acquired in the laboratory.
We have released SAG-QC, which makes the single-cell genome data high accuracy by removing environmental contamination, and ccSAG, which combine multiple single-cell genome data to create a high-quality genome data so far. Currently, we are processing the research to clarify the characteristics of symbiotic microorganisms based on evolutionary genomics by using genome data of microorganisms, and to identify disease-related genes from spatial transcriptome data using new algorithms.
We aim to elucidate biology from various perspectives, by integrating the data obtained in our laboratory with information analysis technologies covering a wide range of fields, such as metagenomes・single-cell genomes・single-cell transcriptomes.
